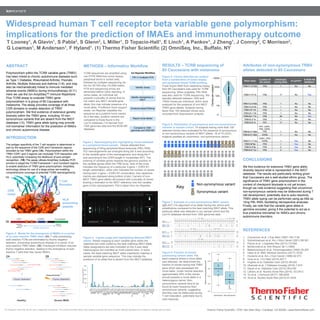
Widespread human T cell receptor beta variable gene polymorphism: implications for the prediction of IMAEs and immunotherapy outcome
- 1. Thermo Fisher Scientific • 5791 Van Allen Way • Carlsbad, CA 92008 • www.thermofisher.comFor Research Use Only. Not for use in diagnostic procedures. The content provided herein may relate to products that have not been officially released and is subject to change without notice. T Looney1, A Glavin1, S Pabla2, S Glenn2, L Miller1, D Topacio-Hall1, E Linch1, A Pankov1, J Zheng1, J Conroy2, C Morrison2, G Lowman1, M Andersen1, F Hyland1. (1) Thermo Fisher Scientific (2) OmniSeq, Inc., Buffalo, NY Widespread human T cell receptor beta variable gene polymorphism: implications for the prediction of IMAEs and immunotherapy outcome METHODS – Informatics Workflow Figure 4. V-gene usage plot highlighting detected IMGT alleles. Reads mapping to each variable gene within the repertoire are color-coded by the best matching IMGT allele. Allele designations are also indicated on the X-axis label. Heterozygous loci manifest as multi-colored bars. In some cases, the best-matching IMGT allele imperfectly matches a sample variable gene sequence. This may indicate the presence of an allele that is absent from the IMGT database. Figure 7. Example of a non-synonymous IMGT variant. IgBLAST (14) alignment of an allele having two amino acid substitutions compared to the best matching IMGT allele. This particular allele was detected in our sample cohort and the Lym1K database derived from 1000 genomes data. Figure 5. Clones detected per subject from a combination of tumor biopsy and peripheral blood TCRB profiling. Fresh frozen RNA from melanoma biopsy from 85 Caucasians was used for TCRB sequencing. When available, PBL RNA was also used for TCRB sequencing. We typically detected between 1000 and 10000 clones per individual, which were analyzed for the presence of non-IMGT TRBV gene alleles. Subjects having fewer than 100 detected clones were excluded from downstream analysis. ABSTRACT Polymorphism within the TCRB variable gene (TRBV) has been linked to chronic autoimmune diseases such as Type 1 Diabetes, Rheumatoid Arthritis, Psoriatic Arthritis, Multiple Sclerosis and Asthma (1-8), and may also be mechanistically linked to immune mediated adverse events (IMAEs) during immunotherapy (9-11). Here we use the Ion-AmpliSeq™ Immune Repertoire Plus TCRB assay to evaluate TRBV gene polymorphism in a group of 85 Caucasians with melanoma. The assay provides coverage of all three CDR domains to enable detection of TRBV polymorphism. We find evidence of extensive genetic diversity within the TRBV gene, including 15 non- synonymous variants that are absent from the IMGT database (12). TRBV gene allele typing may provide rich biomarker information for the prediction of IMAEs and chronic autoimmune disease. INTRODUCTION The antigen specificity of the T cell receptor is determined in part by the sequence of the CDR and Framework regions encoded by the TRBV gene (1A). Polymorphism within the TRBV CDR1 and 2 regions can modulate TCR interaction with HLA, potentially increasing the likelihood of auto-antigen recognition. (1B) The assay utilizes AmpliSeq multiplex PCR primers to target the TCRB Framework 1 and Constant regions to enable detection of TRBV gene polymorphism. AmpliSeq technology allows for use of a large primer set enabling comprehensive coverage of potential TCRB rearrangements. FR1 CDR3 Constant CONCLUSIONS We find evidence for extensive TRBV gene allelic diversity beyond what is represented in the IMGT database. The results are particularly striking given that Caucasians are a well-studied ethnic group. The significance of TRBV gene polymorphism in the context of checkpoint blockade is not yet known, though we note evidence suggesting that uncommon non-synonymous variants may be disfavored during T cell development, potentially due to auto-reactivity. TRBV allele typing can be performed using as little as 10ng PBL RNA, facilitating retrospective analyses. Finally, we note that the variable gene allele is germline encoded, giving it the potential to act as a true predictive biomarker for IMAEs and chronic autoimmune disorders. REFERENCES 1. Concannon et al. J Exp Med (1987) 165:1130 2. Subrahmanyan et al. Am J Hum Genet (2001) 69:381 3. Pierce et al. J Diabetes Res (2013) 737485 4. McDermott et al. Arth Rheum 38:1 (1995) 5. Maksmoyowych et al. Immunogenetics (1992) 35:257 6. Uebe et al. BMC Medical Genetics (2017) 18:92 7. Hockertz et al. Am J Hum Genet (1998) 62:373 8. Gras et al. J Ex Med (2010) 207:7 9. Hughes et al. Diabetes Care (2015) 38:e55 10. Okamoto et al. J Diabetes Investig (2016) 7:915 11. Gaudi et al. Diabetes Care (2015) 38:e182 12. Lefranc et al. Nucleic Acids Res (2015). 43:D413 13. Yu et al. J Immunol (2017) 198:2202 14. Ye et al. Nucleic Acids Res (2013) 41:W34Thymic Selection Severe IMAE Auto-reactive CDR3 Allele name Location of AA variant Individuals having allele In Lym1k? In NCBI NR database? TRBV11-2*32k FR3 1 No No TRBV11-3*1k FR2 17 No Yes TRBV12-4*46k FR2 1 No No TRBV12-5*4k FR2 1 No No TRBV19*17k FR2 1 No No TRBV23-1*2k FR3 1 No No TRBV24-1*1k FR2 39 No Yes TRBV5-3*1k FR2 1 No No TRBV5-8*1k FR1 17 No No TRBV6-2*156k FR1, CDR1, FR2, CDR2, FR3 1 No No TRBV6-5*106k CDR2 1 No No TRBV11-1*11p CDR1, FR2/ CDR2 1 Yes No TRBV30*6p FR3 1 Yes No TRBV5-5*9p FR3 2 Yes No TRBV5-6*11p FR3 3 Yes No Non-synonymous variant Synonymous variant Key 12345 1 100000 10000 1000 100 10 Synonymous Non-synonymous 0.0 0.2 0.4 0.6 0.8 1.0 fractionofclonespossessingvariant Figure 6. Distribution of synonymous and non-synonymous variants in sample cohort. 74 subjects having more than 100 detected clones were evaluated for the presence of synonymous or non-synonymous variants of IMGT alleles. 16 of 74 (22%) subjects possess an uncommon, non-synonymous variant. Figure 8. Fraction of clones possessing variant allele. For each instance where a novel allele was detected, we determined the fraction of clones having that TRBV gene which also possessed the novel allele. Under neutral selection, approximately 50% of the clones should possess a novel allele in a heterozygous carrier. Non- synonymous variants tend to be found at lower frequency than synonymous variants, suggesting that they may be disfavored during T cell maturation, potentially due to auto-reactivity. Auto-reactive TRBV Thymic Selection Healthy Checkpoint blockade Figure 2. Model for the emergence of IMAEs in a carrier of an auto-reactive TRBV allele (2A) T cells possessing auto-reactive TCRs are eliminated by thymic negative selection, preventing autoimmune disease in a carrier of an auto-reactive TRBV allele. (2B) Checkpoint inhibition reduces thymic negative selection, leading to the emergence of auto- reactive T cells that may cause IMAEs. Identify mismatches to IMGT Evidence-based filtering Identify clones FR1-C multiplex PCR Report novel alleles Compare to 1000 genomes and NCBI NR Ion Reporter WorkflowTCRB sequences are amplified using non-FFPE RNA from tumor biopsy, peripheral blood or sorted cells, followed by multiplex sequencing via the Ion S5 530 chip (15-20M reads). PCR and sequencing errors are eliminated before clone reporting. In some cases, an individual will possess a plurality of clones that do not match any IMGT variable gene allele; this may indicate presence of a novel allele. If sufficient clone support exists, Ion Reporter classifies the sequence as a putative novel variant. As a last step, putative variants are compared to those found in the Lym1k database (13) derived from 1000 genomes data and the NCBI NR database. Figure 3. Spectratyping plot highlighting clones detected in a peripheral blood sample. Clones detected from sequencing of 50ng peripheral blood leukocyte (PBL) RNA. VDJ rearrangements are arranged along the X-axis according to the variable gene of the rearrangement, and along the Y- axis according to the CDR3 length in nucleotides (NT). The ordering of variable genes respects the genomic position of the variable genes within the TRB locus. Size of the circle indicates the frequency of a particular V-gene + CDR3 NT length combination, while color indicates the number of clones having each V-gene + CDR3 NT combination. Key repertoire metrics are displayed along bottom of plot. Carriers of non- IMGT TRBV gene alleles will present with a plurality of clones sharing a mismatch to the IMGT reference over the variable gene of the rearrangement. Plot is taken from Ion Reporter. RESULTS – TCRB sequencing of 85 Caucasians with melanoma Attributes of non-synonymous TRBV alleles detected in 85 Caucasians ● 0 20 40 60 80 TCRB V−gene usage and number of clones for B501175_−_PBMC_Leukocyte_−_50ng_RNA_v1_20170811155825372 CDR3length(nt) 1 787 1574 Number of Clones 2112118 readsClone Shannon Diversity: 11.7485 Clone Evenness: 0.742 58431 clones Mean CDR3 NT length: 37.4392 +/− 5.311 V−gene Shannon Diversity: 4.3052 V−CDR3 Frequency 0.10 0.05 ●0.01 ●0.005 ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● TRBV1 TRBV2 TRBV3−1 TRBV4−1 TRBV5−1 TRBV6−1 TRBV7−1 TRBV4−2 TRBV6−2 TRBV3−2 TRBV4−3 TRBV6−3 TRBV7−2 TRBV6−4 TRBV7−3 TRBV5−3 TRBV9 TRBV10−1 TRBV11−1 TRBV12−1 TRBV10−2 TRBV11−2 TRBV12−2 TRBV6−5 TRBV7−4 TRBV5−4 TRBV6−6 TRBV5−5 TRBV6−7 TRBV7−6 TRBV5−6 TRBV6−8 TRBV7−7 TRBV5−7 TRBV6−9 TRBV7−8 TRBV5−8 TRBV7−9 TRBV13 TRBV10−3 TRBV11−3 TRBV12−3 TRBV12−4 TRBV12−5 TRBV14 TRBV15 TRBV16 TRBV17 TRBV18 TRBV19 TRBV20−1 TRBV21−1 TRBV23−1 TRBV24−1 TRBV25−1 TRBV26 TRBV27 TRBV28 TRBV29−1 TRBV30 Clonespersubject CDR loops T Cell Receptor HLA presenting peptide Variable Diversity Joining Constant N1 N2 A. Adult IGH orTCRBeta chain rearrangement In adult B andT cells, the process ofVDJ rearrangement very often involves exonucleotide chewback ofVDJ genes and the addition of non-templated bases, forming N1 and N2 regions in the B cell receptor heavy chain CDR3 and theT cell receptor Beta chain CDR3. These processes vastly increase IGH andTCRB CDR3 diversity. Variable Diversity Joining Constant B. Fetal IGH orTCRBeta chain rearrangement In the fetus, the process ofVDJ rearrangement often occurs without exonucleotide chewback ofVDJ genes and addition of non-templated bases, resulting in a restricted IGH andTCRB CDR3 repertoire that is distinct from the adult repertoire. These structural differences can be used to distinguish fetal B andT cell CDR3 receptors from maternal B andT cell CDR3 receptors in cell free DNA present in maternal peripheral blood. In this way, fetal B andT cell health and development may be monitored in a non-invasive manner. Figure 1. Structural differences between fetal and adult B andT cell receptors ~330bp amplicon 1A 1B 2A 2B TRBV1 TRBV2*01/02 TRBV3−1*01 TRBV4−1*01 TRBV5−1*01 TRBV6−1*01 TRBV7−1 TRBV4−2*01 TRBV6−2*01 TRBV3−2 TRBV4−3*01 TRBV6−3 TRBV7−2*01/02 TRBV6−4*01/02 TRBV7−3*01/03 TRBV5−3*01/02 TRBV9*01/02 TRBV10−1*02 TRBV11−1*01 TRBV12−1 TRBV10−2*01 TRBV11−2*03 TRBV12−2 TRBV6−5*01 TRBV7−4*01 TRBV5−4*01 TRBV6−6*02/01 TRBV5−5*01/02 TRBV6−7*01 TRBV7−6*01 TRBV5−6*01 TRBV6−8*01 TRBV7−7*01 TRBV5−7*01 TRBV6−9*01 TRBV7−8*01 TRBV5−8*01 TRBV7−9*01 TRBV13*01 TRBV10−3*02/03/01 TRBV11−3*01 TRBV12−3*01 TRBV12−4*01 TRBV12−5*01 TRBV14*01/02 TRBV15*02 TRBV16*01 TRBV17 TRBV18*01 TRBV19*01 TRBV20−1*01/05/02 TRBV21−1 TRBV23−1*01 TRBV24−1*01 TRBV25−1*01 TRBV26 TRBV27*01 TRBV28*01 TRBV29−1*01 TRBV30 V−gene usage, colored by allele for B501175_−_PBMC_Leukocyte_−_50ng_RNA_v1_20170811155825372 readshavingv−gene(thousands) 0 100 200 300 400 500
